Creating phylogenetic trees from dna sequences answer key – Welcome to the authoritative guide on creating phylogenetic trees from DNA sequences, where we delve into the intricacies of evolutionary studies and unlock the secrets of genetic relationships. As we embark on this journey, we will explore the fundamental concepts of phylogenetic tree construction, unravel the complexities of DNA sequence alignment, and delve into the algorithms that transform raw data into branching diagrams that illuminate the history of life on Earth.
The content of the second paragraph that provides descriptive and clear information about the topic
Understanding Phylogenetic Tree Construction
Phylogenetic trees are graphical representations of evolutionary relationships among organisms. They depict the branching patterns and common ancestry of species, providing insights into their genetic diversity and evolutionary history.
Phylogenetic trees are constructed using various methods, including:
- Maximum parsimony: Assumes that the tree with the fewest evolutionary changes is the most likely.
- Neighbor-joining: Uses pairwise distances between sequences to infer evolutionary relationships.
- Bayesian inference: Considers statistical models and probability to estimate tree topologies and branch lengths.
DNA Sequence Alignment: Creating Phylogenetic Trees From Dna Sequences Answer Key
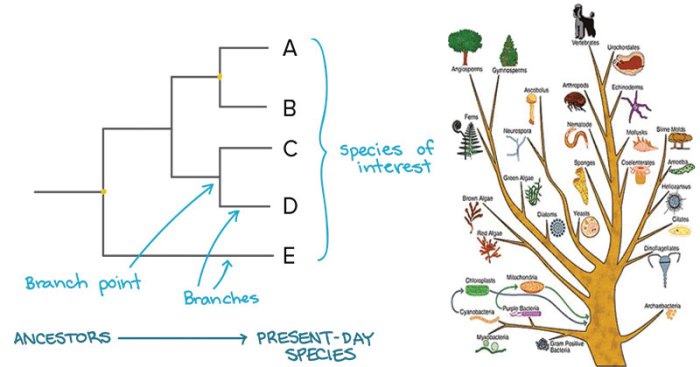
DNA sequence alignment is the process of arranging DNA sequences from different organisms in a way that highlights their similarities and differences. It is crucial for phylogenetic analysis as it allows researchers to compare homologous regions and identify conserved and variable sites.
Methods for DNA sequence alignment include:
- Pairwise alignment: Aligns two sequences at a time.
- Multiple sequence alignment: Aligns multiple sequences simultaneously, considering their evolutionary relationships.
Challenges in DNA sequence alignment include handling gaps and insertions/deletions, considering codon structure, and accounting for evolutionary distance.
Tree Building Algorithms
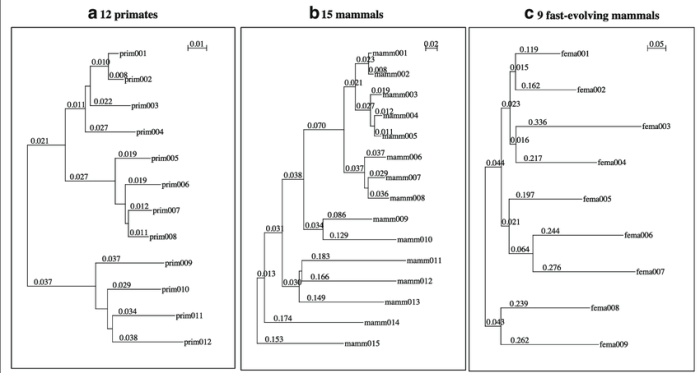
Various algorithms are used to construct phylogenetic trees from DNA sequences. Each algorithm has its advantages and disadvantages:
- Maximum parsimony: Simple and computationally efficient, but may not always find the most accurate tree.
- Neighbor-joining: Fast and widely used, but assumes a constant rate of evolution.
- Bayesian inference: Considers evolutionary models and uncertainty, but computationally intensive.
Software and online tools for tree building include MEGA, PAUP*, and MrBayes.
Tree Evaluation and Interpretation
Evaluating the accuracy and reliability of phylogenetic trees is crucial. Criteria include:
- Tree topology: The branching patterns and relationships between taxa.
- Branch lengths: The evolutionary distances between taxa.
Methods for tree evaluation include:
- Bootstrapping: Resampling data to assess the stability of tree branches.
- Consensus trees: Combining multiple trees to represent uncertainty.
Applications of Phylogenetic Trees

Phylogenetic trees are used in various fields, including:
- Evolutionary biology: Understanding the evolutionary history and relationships among species.
- Systematics: Classifying and organizing organisms based on their evolutionary relationships.
- Medicine: Studying the evolution of pathogens, drug resistance, and genetic diseases.
Limitations and biases in phylogenetic tree interpretation include the accuracy of DNA sequences, the choice of tree-building algorithm, and the evolutionary models used.
Clarifying Questions
What is the significance of phylogenetic trees in evolutionary studies?
Phylogenetic trees provide a visual representation of the evolutionary relationships among different species, allowing scientists to infer their common ancestry and understand the patterns of diversification over time.
How does DNA sequence alignment contribute to phylogenetic tree construction?
DNA sequence alignment is crucial for identifying homologous regions in DNA sequences, which are essential for inferring evolutionary relationships. By aligning sequences, researchers can identify conserved and variable regions that provide insights into the genetic changes that have occurred over time.
What are the advantages and disadvantages of different tree building algorithms?
Different tree building algorithms have their own advantages and disadvantages. Maximum parsimony assumes that the simplest tree with the fewest evolutionary changes is the most likely, while neighbor-joining considers the distances between sequences. Bayesian inference uses statistical models to estimate the probability of different tree topologies.
How can we evaluate the accuracy and reliability of phylogenetic trees?
The accuracy and reliability of phylogenetic trees can be evaluated using criteria such as bootstrap values, which indicate the support for different branches in the tree. Consensus trees can also be used to identify areas of agreement and disagreement among different tree building methods.